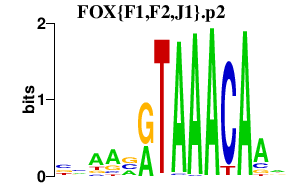
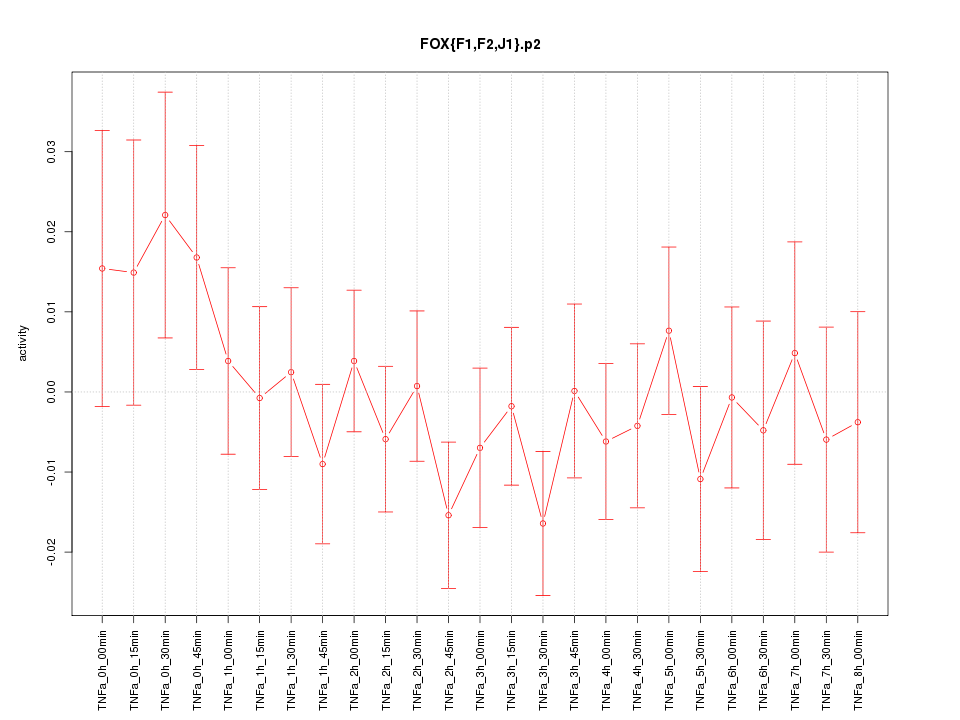
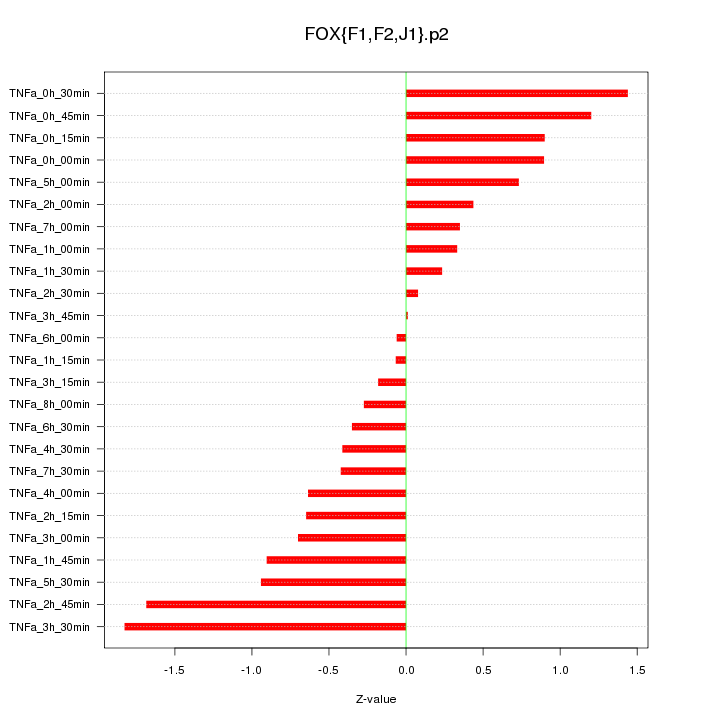
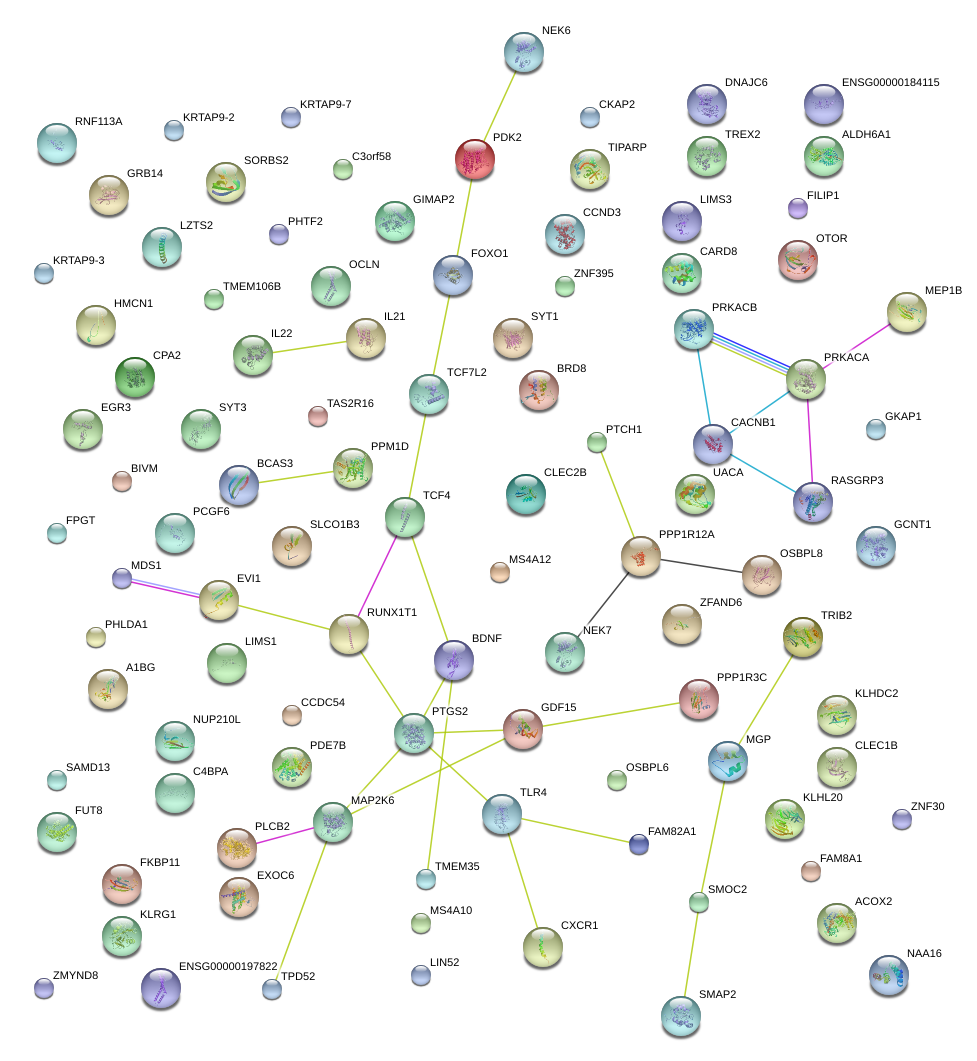
|
chr20_-_45976626
|
2.063
|
|
ZMYND8
|
zinc finger, MYND-type containing 8
|
|
chr8_-_93075190
|
1.755
|
NM_004349
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr6_+_136172802
|
1.691
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr2_-_165424993
|
1.672
|
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr16_-_86542463
|
1.606
|
|
LOC400550
|
uncharacterized LOC400550
|
|
chr8_-_28243941
|
1.502
|
NM_018660
|
ZNF395
|
zinc finger protein 395
|
|
chr8_-_22550708
|
1.466
|
NM_004430
|
EGR3
|
early growth response 3
|
|
chr3_+_143692166
|
1.345
|
NM_001134470
|
C3orf58
|
chromosome 3 open reading frame 58
|
|
chr8_-_93107442
|
1.281
|
NM_001198629
NM_001198630
NM_001198632
NM_175635
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_-_154127591
|
1.244
|
NM_001159484
NM_207308
|
NUP210L
|
nucleoporin 210kDa-like
|
|
chr3_-_168865521
|
1.194
|
NM_001164000
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr11_-_27723061
|
1.180
|
NM_170733
|
BDNF
|
brain-derived neurotrophic factor
|
|
chr9_+_120466453
|
1.156
|
NM_138554
|
TLR4
|
toll-like receptor 4
|
|
chr9_+_79093256
|
1.101
|
NM_001097635
|
GCNT1
|
glucosaminyl (N-acetyl) transferase 1, core 2
|
|
chr9_-_86432546
|
1.096
|
NM_001135953
NM_025211
|
GKAP1
|
G kinase anchoring protein 1
|
|
chr7_+_129906702
|
1.082
|
NM_001869
|
CPA2
|
carboxypeptidase A2 (pancreatic)
|
|
chr9_+_120466607
|
1.059
|
|
TLR4
|
toll-like receptor 4
|
|
chr12_-_15038724
|
1.038
|
NM_000900
NM_001190839
|
MGP
|
matrix Gla protein
|
|
chr2_+_12856813
|
0.993
|
NM_021643
|
TRIB2
|
tribbles homolog 2 (Drosophila)
|
|
chr15_+_80390757
|
0.985
|
NM_001242919
|
ZFAND6
|
zinc finger, AN1-type domain 6
|
|
chr9_+_120466625
|
0.935
|
|
TLR4
|
toll-like receptor 4
|
|
chr12_-_80307690
|
0.925
|
NM_001143886
|
PPP1R12A
|
protein phosphatase 1, regulatory subunit 12A
|
|
chr6_-_76203256
|
0.920
|
NM_015687
|
FILIP1
|
filamin A interacting protein 1
|
|
chr3_+_156392369
|
0.917
|
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr9_-_3223007
|
0.914
|
|
|
|
|
chr19_+_18496966
|
0.907
|
NM_004864
|
GDF15
|
growth differentiation factor 15
|
|
chr22_-_17747520
|
0.905
|
|
CECR3
|
cat eye syndrome chromosome region, candidate 3 (non-protein coding)
|
|
chr8_-_93107643
|
0.889
|
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_65730423
|
0.884
|
NM_014787
|
DNAJC6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr12_-_10021932
|
0.883
|
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr8_-_93115453
|
0.880
|
NM_001198633
NM_175634
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr1_+_22351984
|
0.879
|
|
LINC00339
|
long intergenic non-protein coding RNA 339
|
|
chr4_-_186732047
|
0.868
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr3_+_156392204
|
0.835
|
NM_015508
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr3_-_168864066
|
0.832
|
NM_001105077
NM_001163999
NM_005241
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr13_+_41885340
|
0.819
|
NM_001110798
NM_018527
NM_024561
|
NAA16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit
|
|
chr17_+_39382878
|
0.817
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr7_+_12250818
|
0.808
|
NM_001134232
NM_018374
|
TMEM106B
|
transmembrane protein 106B
|
|
chr5_-_997408
|
0.798
|
NM_001242737
|
LOC100506688
|
uncharacterized LOC100506688
|
|
chr17_+_67410825
|
0.797
|
NM_002758
|
MAP2K6
|
mitogen-activated protein kinase kinase 6
|
|
chr2_+_33661392
|
0.757
|
NM_170672
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated)
|
|
chr4_-_186732208
|
0.726
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr1_+_185703470
|
0.725
|
NM_031935
|
HMCN1
|
hemicentin 1
|
|
chr19_+_35417806
|
0.686
|
NM_001099437
NM_001099438
NM_194325
|
ZNF30
|
zinc finger protein 30
|
|
chr8_-_80993009
|
0.686
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr12_+_79258588
|
0.681
|
|
SYT1
|
synaptotagmin I
|
|
chr14_-_74551159
|
0.677
|
NM_005589
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr15_-_70994608
|
0.672
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr1_+_84630575
|
0.663
|
NM_001242860
NM_001242861
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr1_+_84767288
|
0.656
|
NM_001134663
|
SAMD13
|
sterile alpha motif domain containing 13
|
|
chr3_+_107096187
|
0.648
|
NM_032600
|
CCDC54
|
coiled-coil domain containing 54
|
|
chr9_+_127054246
|
0.639
|
NM_001166170
NM_001166171
|
NEK6
|
NIMA (never in mitosis gene a)-related kinase 6
|
|
chr10_+_94594469
|
0.634
|
NM_001013848
|
EXOC6
|
exocyst complex component 6
|
|
chr14_-_74551073
|
0.628
|
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1
|
|
chr17_+_58677603
|
0.609
|
|
PPM1D
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D
|
|
chr9_-_98268866
|
0.608
|
|
PTCH1
|
patched 1
|
|
chr13_+_103459495
|
0.586
|
NM_001204425
|
BIVM-ERCC5
BIVM
|
BIVM-ERCC5 readthrough
basic, immunoglobulin-like variable motif containing
|
|
chr12_-_10022457
|
0.584
|
NM_005127
|
CLEC2B
|
C-type lectin domain family 2, member B
|
|
chr17_+_48172100
|
0.582
|
NM_001199898
|
PDK2
|
pyruvate dehydrogenase kinase, isozyme 2
|
|
chr11_+_60552820
|
0.575
|
NM_206893
|
MS4A10
|
membrane-spanning 4-domains, subfamily A, member 10
|
|
chr17_-_5138049
|
0.564
|
NM_207103
|
SCIMP
|
SLP adaptor and CSK interacting membrane protein
|
|
chr12_+_9142136
|
0.563
|
NM_005810
|
KLRG1
|
killer cell lectin-like receptor subfamily G, member 1
|
|
chr15_-_40600118
|
0.558
|
NM_004573
|
PLCB2
|
phospholipase C, beta 2
|
|
chr17_+_58755158
|
0.558
|
NM_001099432
NM_017679
|
BCAS3
|
breast carcinoma amplified sequence 3
|
|
chr20_+_16728997
|
0.558
|
NM_020157
|
OTOR
|
otoraplin
|
|
chr12_-_76425375
|
0.555
|
NM_007350
|
PHLDA1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr5_+_68788118
|
0.555
|
NM_002538
|
OCLN
|
occludin
|
|
chrX_+_100333835
|
0.551
|
NM_021637
|
TMEM35
|
transmembrane protein 35
|
|
chr14_+_65877309
|
0.551
|
NM_004480
NM_178155
NM_178156
|
FUT8
|
fucosyltransferase 8 (alpha (1,6) fucosyltransferase)
|
|
chr19_-_48752921
|
0.550
|
NM_001184901
NM_001184903
NM_014959
|
CARD8
|
caspase recruitment domain family, member 8
|
|
chr10_-_105110823
|
0.550
|
|
PCGF6
|
polycomb group ring finger 6
|
|
chr3_-_168864325
|
0.549
|
NM_001105078
|
MECOM
|
MDS1 and EVI1 complex locus
|
|
chr2_+_179184970
|
0.548
|
NM_145739
|
OSBPL6
|
oxysterol binding protein-like 6
|
|
chr1_+_207277577
|
0.545
|
NM_000715
|
C4BPA
|
complement component 4 binding protein, alpha
|
|
chr10_-_105110890
|
0.542
|
NM_001011663
NM_032154
|
PCGF6
|
polycomb group ring finger 6
|
|
chr14_+_74551655
|
0.532
|
NM_001024674
|
LIN52
|
lin-52 homolog (C. elegans)
|
|
chr13_-_41240607
|
0.527
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr10_-_93392744
|
0.524
|
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr10_+_102756829
|
0.520
|
NM_032429
|
LZTS2
|
leucine zipper, putative tumor suppressor 2
|
|
chr7_-_122635753
|
0.517
|
NM_016945
|
TAS2R16
|
taste receptor, type 2, member 16
|
|
chr15_-_40600025
|
0.512
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr13_+_53035227
|
0.511
|
|
CKAP2
|
cytoskeleton associated protein 2
|
|
chr12_+_20963637
|
0.507
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr1_+_173684061
|
0.507
|
NM_014458
|
KLHL20
|
kelch-like 20 (Drosophila)
|
|
chr2_+_109223600
|
0.500
|
NM_001193482
|
LIMS1
|
LIM and senescent cell antigen-like domains 1
|
|
chr19_-_58864820
|
0.499
|
NM_130786
|
A1BG
|
alpha-1-B glycoprotein
|
|
chr2_+_38155901
|
0.499
|
NM_001170792
|
FAM82A1
|
family with sequence similarity 82, member A1
|
|
chr1_+_74701070
|
0.497
|
NM_015978
|
TNNI3K
|
TNNI3 interacting kinase
|
|
chr4_-_123542165
|
0.495
|
NM_001207006
NM_021803
|
IL21
|
interleukin 21
|
|
chr7_+_150382783
|
0.492
|
NM_015660
|
GIMAP2
|
GTPase, IMAP family member 2
|
|
chr6_-_42016383
|
0.492
|
|
CCND3
|
cyclin D3
|
|
chr19_-_51141301
|
0.491
|
NM_032298
|
SYT3
|
synaptotagmin III
|
|
chr18_+_29769986
|
0.490
|
NM_005925
|
MEP1B
|
meprin A, beta
|
|
chr6_+_17600504
|
0.487
|
NM_016255
|
FAM8A1
|
family with sequence similarity 8, member A1
|
|
chr7_+_77469446
|
0.486
|
NM_001127357
NM_001127359
NM_020432
|
PHTF2
|
putative homeodomain transcription factor 2
|
|
chr2_-_165478357
|
0.484
|
NM_004490
|
GRB14
|
growth factor receptor-bound protein 14
|
|
chr18_-_53068755
|
0.483
|
|
TCF4
|
transcription factor 4
|
|
chr10_-_93392857
|
0.479
|
NM_005398
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C
|
|
chr3_-_58522873
|
0.478
|
NM_003500
|
ACOX2
|
acyl-CoA oxidase 2, branched chain
|
|
chr1_+_40862471
|
0.477
|
NM_001198980
|
SMAP2
|
small ArfGAP2
|
|
chr1_-_186649421
|
0.471
|
NM_000963
|
PTGS2
|
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase)
|
|
chr14_+_50234783
|
0.468
|
NM_014315
|
KLHDC2
|
kelch domain containing 2
|
|
chr2_-_219031646
|
0.465
|
NM_000634
|
CXCR1
|
chemokine (C-X-C motif) receptor 1
|
|
chr8_+_74903563
|
0.460
|
NM_001195797
NM_015364
|
LY96
|
lymphocyte antigen 96
|
|
chr3_+_98451537
|
0.458
|
NM_006100
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6
|
|
chr2_+_86947387
|
0.458
|
NM_022780
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr14_+_50234907
|
0.455
|
|
KLHDC2
|
kelch domain containing 2
|
|
chr10_-_50732142
|
0.453
|
NM_170753
|
PGBD3
|
piggyBac transposable element derived 3
|
|
chr20_+_45338125
|
0.451
|
NM_030777
|
SLC2A10
|
solute carrier family 2 (facilitated glucose transporter), member 10
|
|
chr2_+_1417232
|
0.451
|
NM_000547
NM_001206744
NM_001206745
NM_175719
|
TPO
|
thyroid peroxidase
|
|
chr17_-_7082559
|
0.449
|
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr1_+_246729818
|
0.448
|
|
CNST
|
consortin, connexin sorting protein
|
|
chr1_-_57431620
|
0.447
|
NM_000066
|
C8B
|
complement component 8, beta polypeptide
|
|
chr19_+_18111923
|
0.447
|
NM_001025604
|
ARRDC2
|
arrestin domain containing 2
|
|
chr5_-_39424930
|
0.446
|
|
DAB2
|
disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila)
|
|
chr9_-_35812143
|
0.445
|
NM_001039592
NM_172312
|
SPAG8
|
sperm associated antigen 8
|
|
chr9_-_124984018
|
0.438
|
NM_001242335
|
LHX6
|
LIM homeobox 6
|
|
chr5_+_68800003
|
0.435
|
NM_001205255
|
OCLN
|
occludin
|
|
chr11_+_113931228
|
0.430
|
NM_001018011
|
ZBTB16
|
zinc finger and BTB domain containing 16
|
|
chr17_-_7082654
|
0.428
|
NM_001197216
NM_001671
|
ASGR1
|
asialoglycoprotein receptor 1
|
|
chr14_+_100563817
|
0.425
|
|
EVL
|
Enah/Vasp-like
|
|
chr10_-_105110844
|
0.421
|
|
PCGF6
|
polycomb group ring finger 6
|
|
chr15_-_40599872
|
0.415
|
|
PLCB2
|
phospholipase C, beta 2
|
|
chr17_+_11786218
|
0.412
|
NM_004662
|
DNAH9
|
dynein, axonemal, heavy chain 9
|
|
chr10_-_49482912
|
0.412
|
NM_001018071
|
FRMPD2
|
FERM and PDZ domain containing 2
|
|
chr10_+_5136589
|
0.411
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr9_+_134000939
|
0.410
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr3_+_23987514
|
0.409
|
NM_001145425
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2
|
|
chr2_-_87248942
|
0.407
|
NM_002665
NM_001032392
|
PLGLB2
PLGLB1
|
plasminogen-like B2
plasminogen-like B1
|
|
chr3_+_132036210
|
0.406
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr2_-_170430312
|
0.406
|
NM_024622
|
FASTKD1
|
FAST kinase domains 1
|
|
chr6_-_42016609
|
0.406
|
NM_001136017
NM_001136126
|
CCND3
|
cyclin D3
|
|
chr8_+_79428335
|
0.404
|
NM_006823
NM_181839
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha
|
|
chr15_+_67000929
|
0.403
|
NM_001142861
|
SMAD6
|
SMAD family member 6
|
|
chr12_-_86229981
|
0.401
|
|
RASSF9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9
|
|
chr17_+_3377346
|
0.399
|
NM_001128085
|
ASPA
|
aspartoacylase
|
|
chr10_-_125853102
|
0.399
|
|
CHST15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr5_+_140579182
|
0.398
|
NM_018931
|
PCDHB11
|
protocadherin beta 11
|
|
chr2_+_86947429
|
0.396
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr2_+_220042948
|
0.394
|
|
FAM134A
|
family with sequence similarity 134, member A
|
|
chr8_-_110655418
|
0.394
|
NM_001099756
|
SYBU
|
syntabulin (syntaxin-interacting)
|
|
chr5_+_59784004
|
0.394
|
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding)
|
|
chr1_+_84630374
|
0.390
|
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr6_-_112575880
|
0.389
|
|
LAMA4
|
laminin, alpha 4
|
|
chr8_-_93107881
|
0.389
|
NM_001198625
NM_001198626
NM_001198627
NM_001198631
NM_001198634
NM_001198679
|
RUNX1T1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr18_+_61575203
|
0.389
|
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10
|
|
chr2_+_175260505
|
0.387
|
|
SCRN3
|
secernin 3
|
|
chr1_-_24469571
|
0.387
|
NM_021258
|
IL22RA1
|
interleukin 22 receptor, alpha 1
|
|
chr2_+_175260455
|
0.387
|
NM_001193528
NM_024583
|
SCRN3
|
secernin 3
|
|
chr10_+_23728197
|
0.384
|
NM_001145373
|
OTUD1
|
OTU domain containing 1
|
|
chr1_+_61542928
|
0.383
|
NM_001145511
|
NFIA
|
nuclear factor I/A
|
|
chr10_+_5136556
|
0.383
|
NM_003739
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr6_+_43027628
|
0.383
|
NM_201523
|
KLC4
|
kinesin light chain 4
|
|
chrX_-_63005212
|
0.379
|
NM_001173479
|
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9
|
|
chr5_-_180076616
|
0.378
|
NM_002020
NM_182925
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr20_-_17539480
|
0.376
|
NM_001161705
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr6_-_160679789
|
0.375
|
NM_003058
|
SLC22A2
|
solute carrier family 22 (organic cation transporter), member 2
|
|
chr2_+_1418179
|
0.374
|
NM_175721
NM_175722
|
TPO
|
thyroid peroxidase
|
|
chr4_-_16900348
|
0.374
|
NM_001130834
NM_001290
|
LDB2
|
LIM domain binding 2
|
|
chr15_+_72978519
|
0.372
|
NM_001252678
NM_033028
|
BBS4
|
Bardet-Biedl syndrome 4
|
|
chr4_+_36285643
|
0.371
|
NM_001170700
|
DTHD1
|
death domain containing 1
|
|
chr10_-_5045967
|
0.370
|
NM_001135241
|
AKR1C2
|
aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III)
|
|
chr10_-_50396380
|
0.367
|
NM_001010863
|
C10orf128
|
chromosome 10 open reading frame 128
|
|
chr1_-_247697140
|
0.367
|
NM_198074
|
OR2C3
|
olfactory receptor, family 2, subfamily C, member 3
|
|
chr5_+_150157507
|
0.366
|
NM_032947
|
C5orf62
|
chromosome 5 open reading frame 62
|
|
chr2_+_86947570
|
0.365
|
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae)
|
|
chr6_-_112575766
|
0.365
|
NM_001105206
NM_001105207
NM_001105208
NM_001105209
NM_002290
|
LAMA4
|
laminin, alpha 4
|
|
chr5_-_180076572
|
0.364
|
|
FLT4
|
fms-related tyrosine kinase 4
|
|
chr13_-_48987652
|
0.364
|
NM_001162498
|
LPAR6
|
lysophosphatidic acid receptor 6
|
|
chr6_-_56407681
|
0.360
|
|
|
|
|
chr4_-_16900256
|
0.359
|
|
LDB2
|
LIM domain binding 2
|
|
chr17_-_39334439
|
0.359
|
NM_033062
|
KRTAP4-2
|
keratin associated protein 4-2
|
|
chr10_+_5136613
|
0.358
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr4_-_16900028
|
0.355
|
|
LDB2
|
LIM domain binding 2
|
|
chr2_+_191301020
|
0.354
|
|
MFSD6
|
major facilitator superfamily domain containing 6
|
|
chr17_+_3379295
|
0.349
|
NM_000049
|
ASPA
|
aspartoacylase
|
|
chr2_-_87303105
|
0.346
|
|
LOC285074
|
anaphase promoting complex subunit 1 pseudogene
|
|
chr5_+_131892556
|
0.345
|
NM_005732
|
RAD50
|
RAD50 homolog (S. cerevisiae)
|
|
chr15_-_65412175
|
0.342
|
|
PDCD7
|
programmed cell death 7
|
|
chr6_+_44244465
|
0.341
|
|
TMEM151B
|
transmembrane protein 151B
|
|
chr17_+_72427535
|
0.339
|
NM_022036
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C
|
|
chr3_-_15839674
|
0.337
|
NM_001195098
|
ANKRD28
|
ankyrin repeat domain 28
|
|
chr1_+_84630014
|
0.337
|
NM_001242857
NM_001242858
NM_001242859
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta
|
|
chr18_-_53071225
|
0.336
|
NM_001243232
|
TCF4
|
transcription factor 4
|
|
chr1_+_82266081
|
0.336
|
NM_012302
|
LPHN2
|
latrophilin 2
|
|
chr1_+_207002221
|
0.334
|
NM_013371
|
IL19
|
interleukin 19
|
|
chr8_-_77912296
|
0.333
|
NM_000318
NM_001079867
|
PEX2
|
peroxisomal biogenesis factor 2
|
|
chr1_+_246729638
|
0.331
|
NM_001139459
NM_152609
|
CNST
|
consortin, connexin sorting protein
|
|
chrX_-_135863502
|
0.331
|
NM_004840
|
ARHGEF6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr5_+_140220811
|
0.330
|
NM_018911
NM_031856
|
PCDHA8
|
protocadherin alpha 8
|
|
chr4_+_155484131
|
0.329
|
NM_001184741
NM_005141
|
FGB
|
fibrinogen beta chain
|
|
chr3_-_183273407
|
0.329
|
NM_130446
|
KLHL6
|
kelch-like 6 (Drosophila)
|
|
chr2_-_142889269
|
0.327
|
NM_018557
|
LRP1B
|
low density lipoprotein receptor-related protein 1B
|
|
chr5_+_54398473
|
0.327
|
NM_006144
|
GZMA
|
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3)
|
|
chr6_+_26501448
|
0.327
|
NM_001732
|
BTN1A1
|
butyrophilin, subfamily 1, member A1
|
|
chr3_+_132316080
|
0.323
|
NM_016557
|
CCRL1
|
chemokine (C-C motif) receptor-like 1
|
|
chr3_-_165555252
|
0.319
|
NM_000055
|
BCHE
|
butyrylcholinesterase
|
|
chr19_+_41699076
|
0.317
|
NM_030622
|
CYP2S1
|
cytochrome P450, family 2, subfamily S, polypeptide 1
|
|
chr14_+_102276125
|
0.316
|
NM_002719
NM_178586
NM_178587
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma
|
|
chr14_-_106714594
|
0.316
|
|
|
|